
 Research Projects
Research Projects
 Rational design of Armadillo Repeat Proteins as a modular recognition system for the sequence-specific binding of peptides
Rational design of Armadillo Repeat Proteins as a modular recognition system for the sequence-specific binding of peptides
The specific recognition of peptide sequences by antibodies plays an important role in cell biology and diagnostic applications but the production of antibodies is time-consuming. In this project in collaboration with the groups of Andreas Plückthun, Markus Grütter and Amedeo Caflish we aim to develop Armadillo repeat proteins to bind extended peptides in a modular way. Therein, each repeat represents one module binding a 'two amino acid'-fragment of a peptide:
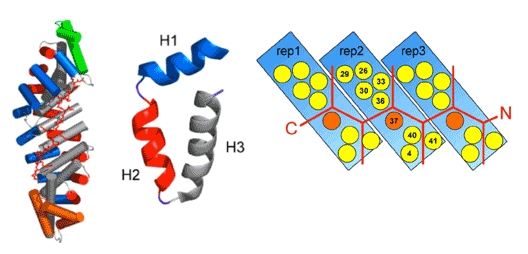
NMR will be used to access i) the structural stability of the developed proteins, ii) to test whether peptides bind to these proteins and iii) to characterize the binding mode of the peptides. This information will be directly used for the design of the proteins.
We have now developed NMR methodology for resonance assignments of these proteins that contain peptide fragments identical in sequence. We have applied it to full-consensus ankyrins repeats and studied their folding properties using hydrogen exchange, chemical shift mapping and backbone dynamics.
![]() Structural Studies of Large Fragments of G-protein coupled receptors
Structural Studies of Large Fragments of G-protein coupled receptors
In an attempt to structurally characterize G-protein coupled receptors we are expressing or synthesizing fragments from the Neuropeptide Y (the so-called Y) receptors. The fragments are produced recombinantly as fusions to insoluble proteins in isotopically labeled form. The fragments are solubilized in phospholipids micelles and structurally characterized by high-resolution NMR spectroscopy. Structural work is supported by measurement of internal backbone dynamics and elucidation of the membrane association topology. Moreover, binding to possible ligands is tested by chemical shift mapping technologies. Here we use both the native ligands, hormones from the NPY family, as well as control peptides in which residues believed to be important for binding are replaced. This work is initiated in order to improve our understanding of the structure of these pharmaceutically highly important receptors, as well as to shed light on the scenario of events that take place during receptor binding.
One system that was recently investigated comprises constructs derived from the Y4 receptor. The N-terminal (extracellular) domain was structurally characterized by NMR . In addition, we tested binding of neurohormones form the NPY family to the domain using chemical shift mapping and surface plasmon resonance. Furthermore, studies incldued a construct that contains the N-terminal domain fused to the first two transmembrane helices. Moreover, we are working on double-TM helix fragments containing either N-TM1-TM2, TM4-TM5 or TM6-TM7.
On another project we are determining structures of fragments from the yeast Ste2p GPCR in collaboration with Fred Naider from the City University of New York and Jeff Becker from Tennessee. Again, we are determining structures of fragments by NMR in phospholipids micelles. Initially, we elucidated the structure of a fragment comprising the seventh transmembrane helix as well as as part of the cytosolic loop. We subsequently determined the structure of a 80 residue polypeptide in LPPG micelles, that contains the first 2 TM helices. Again, we are moving to other and longer fragments.
![]() Recognition of ligands to GPCRs
Recognition of ligands to GPCRs
We have recently been working on the recognition of G-protein coupled receptors by hormones. For our studies we have characterized the neuropeptide Y (NPY)/Y-receptor system. NPY is the most abundant neurohormone in the central nervous system. For binding of NPY to its receptors we have postulated a three-step model, which includes the membrane association as an important step that precedes receptor binding of the ligand.
The conducted analysis comprised the following steps:
- Elucidation of the structure of micelle-bound peptides
- Determination of the membrane-binding topology by 15N,1H correlation spectroscopy utilizing micelle-integrating spinlabels
- Determination of the dynamics of membrane-bound peptides by 15N relaxation
By comparison with the solution structure of NPY we discovered that structural changes are introduced upon binding to the membrane [1]. In particular, residues which are known to mediate receptor contacts become located close to the membrane, water interface in a well defined manner.
We have recently been looking into possible structural determinants for receptor subtype specificity. Firstly, we have characterized the solution structure of [31Ala,32Aib]-NPY, a mutant that selectively binds to the Y-5 receptor and induces increases in food-intake [2]. We discovered larger structural differences in the C-terminal part of the a-helix. Furthermore, we have been looking at structural differences of another Y-5 selective mutant [31Ala,32Pro]-NPY in its membrane-bound form and compared it to wild-type NPY [3].
Presently, we investigate a number of other, subtype-specific ligands in order to verify our postulate. We have also determined the structure and dynamics of the pancreatic polypeptide (PP), a peptide that selectively targets the Y4 receptor subtype[4]. Moreover, we compared NPY and PYY, two pharmacologically closely related peptides, and discovered that similarities in pharmacology are better related to strcutural features of the membrane-bound state[5]. We have also looked into NPY/PP chimaera with interesting biological properties in order to yield a better understanding of receptor subtype selectivity[6]. All those experiments have recently reviewed by us[7].
We are now looking into structures of N terminal fragments of Y receptors and test binding of these constructs to wild-type hormones in order to understand to which extent contacts with the N-terminal domain of the Y receptors may be responsible for receptor recognition and binding.
![]() Models to mimic binding epitopes of GPCRs
Models to mimic binding epitopes of GPCRs
Biosynthetic production of G-protein coupled receptors is a difficult task. The aim of this project is to graft binding epitopes of the Y receptors, which represent receptors of the pharmacologically very important class of hormones of the neuropeptide Y family, onto a rigid scaffold. The use of so-called protein scaffolds has recently been introduced to engineer artificial receptors for prescribed ligands based on a known polypeptide fold. Typically, such scaffolds are built upon units of rigid secondary structure, and loops with intrinsic structural plasticity are targeted for mutagenesis. To be successful the scaffold the scaffold should be conformationally rigid, well producible in bacterial expression systems, especially in order to facilitate biosynthesis of isotopically enriched proteins for NMR studies, be soluble, preferably monomeric, and in a size range that is amenable to high-resolution NMR studies. Furthermore, it needs to possess at least three different loops located on a single contiguous surface (and exposed to the same side of the protein). In view of the requirements described above we have chosen beta-barrel proteins as scaffolds. So far, both soluble beta-barrel proteins from the class of the so-called lipocalins as well as beta-barrel proteins from the outer membrane of E. coli were tested. Grafting of complete loops could be successfully achieved in case of the membrane proteins. Optimization of constructs, testing of their binding of the natural ligands, and characterization of the structure and binding epitopes of these proteins is currently under way.
![]() Cell-surface exposed glycolipids
Cell-surface exposed glycolipids
Cell-cell recognition is an event of prime biological importance in a variety of biological phenomena such as cell migration, organ formation, immune defense and microbial infection. Specificity of the interaction is mediated by various, tissue-dependent, receptors.We have initiated studies to create a functional model for cell-surface exposed carbohydrate units that can be used to investigate interactions with lectins, and which help to increase the complexity of our membrane models. We have started our studies with systems of the following architecture: A lipid anchor is extended by an flexible linker, onto which the carbohydrate portion is coupled. We have recently been been able to show that these systems are properly intergrated into our phospholipid micelles.
We have also investigated binding of Cyanovirin (CNV) to micelle-immobilized 1,2-dimannoside and 1,2 linked trimannoside and could demonstrate that the protein is properly recognized. CVN is a cyanobacterial protein that interacts through high-affinity carbohydrate-mediated interactions with the surface-envelope glycoprotein gp120 from various HIV and SIV strains thereby disrupting the gp120-CD4 interaction and blocking HIV entry. We could also demonstrate that CVN linked to micelles could be released by adding a higher affinity ligand such as the (non-lipidated) trimannoside. By adding lectin domains to proteins a system capable of reversible anchoring proteins onto membranes is created that circumvents problems with adding chemical moieties onto proteins.
In addition we looked at the complex between lipopolysaccharide (LPS) and antimicrobial peptides of the polymyxin type. LPS was labelled and purified, and the glycolipid measured when incorporated in DPC micelles. The data derived from chemical shift mapping and heteronuclear NOEs together with restraints from MD caclulation allowed deriving a good resolution picture of the bimolecular complex.