Site-specific modification of Nucleic Acids
Site-specific modification of Nucleic Acids
The basic concept applied makes use of DNA-templated synthesis. A modified oligonucleotide sequence is used to guide a specifically designed reactive group to the nucleotide in the target sequence that is to be modified. We apply organic synthesis in combination with biochemical methods, e.g. PAGE and enzymatic assays. Additionally, HPLC methods, mass spectrometry, UV, CD, fluorescence, and NMR spectroscopy play major roles. The crucial point of this research is to find a compromise between the conditions required for the chemical reaction leading to the desired modification and the stability of the target molecule, i.e. DNA or RNA.
Gene Cloning, Protein Over-Expression, and Purification
To overcome certain drawbacks associated with the direct isolation of proteins from plant material – plant growth, low yields, inhomogeneous protein batches, proteolytic degradation... – we construct all of our plant MT genes with common polymerase chain reaction (PCR) methods and clone them into suitable vectors for recombinant protein expression in E. coli. This provides us with high quality material in sufficient yield for our further investigations and also opens up a straight forward way to directly manipulate the genetic sequence and introduce specific mutations and alterations when required for our experiments.
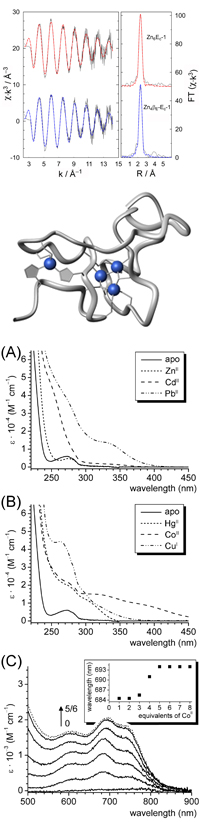
Spectroscopic and Biochemical Characterization of MTs
MTs can be observed with UV-Vis spectroscopy due to the characteristic absorption bands originating from the metal-thiolate clusters. Furthermore, the position and intensity of these so called ligand-to-metal charge transfer (LMCT) bands is indicative for the sort of metal ions bound. Therefore, UV-Vis spectroscopy is always the method of choice for the initial characterization of a new plant MT. Additionally, (magnetic) circular dichroism and fluorescence spectroscopy combined with biochemical methods are applied to obtain information such as the metal ion to protein stoichiometry, the metal ion binding affinity, and also about the probable metal-thiolate cluster organisation. MS measurements complement the characterisation at each stage.
Determination of Metal Ion Binding Affinities and Redox States
Metal ion binding affinities vary in the different plant MTs, especially between the different sub-groups, and also strongly depend on the nature of the coordinated metal ion. Investigations are performed with the afore mentioned spectroscopic techniques, supplemented with e.g. potentiometric measurements and competition assays with metal ion chelators of known characteristics. Knowledge of the redox state is crucial when evaluating metal ion binding, both of the metal ion as well as of the protein itself, specifically of the oxidation sensitive cysteine residues involved in metal ion binding.
Analysis of Protein Folding
The metal-thiolate clusters dictate the three-dimensional structures of MTs, which are largely unstructured in the metal-free state. Accordingly, the study of MT folding upon metal ion coordination is of great interest and can be studied with NMR, fluorescence spectroscopy or dynamic light scattering.
Structure determination of MTs by X-ray and NMR
One of the main goals is the determination of the three-dimensional structures with single crystal X-ray crystallography and NMR spectroscopy. Excellent facilities are available at the Department of Chemistry and the Institute of Biochemistry, and the Swiss Light Source, a third-generation synchrotron facility, is located close to Zurich. We were already able to determine the very first two structures of plant MTs and continue working on the other three plant MT subfamilies known so far.