Software Development
The processing and analysis of microscopy and NMR data in the scope of RNA-metal ion interaction studies require the use of very diverse computing tools. To characterize experimental evidences and increase the significance of our findings, we dedicate a non-negligible part of our research to software development. In this domain, we aim to address three problems:
- the handling and analysis of complex single molecule FRET (sm-FRET) data,
- the management of existing information about metal ions in nucleic acid structures,
- the characterization of metal ion affinity to RNA in NMR experiments.
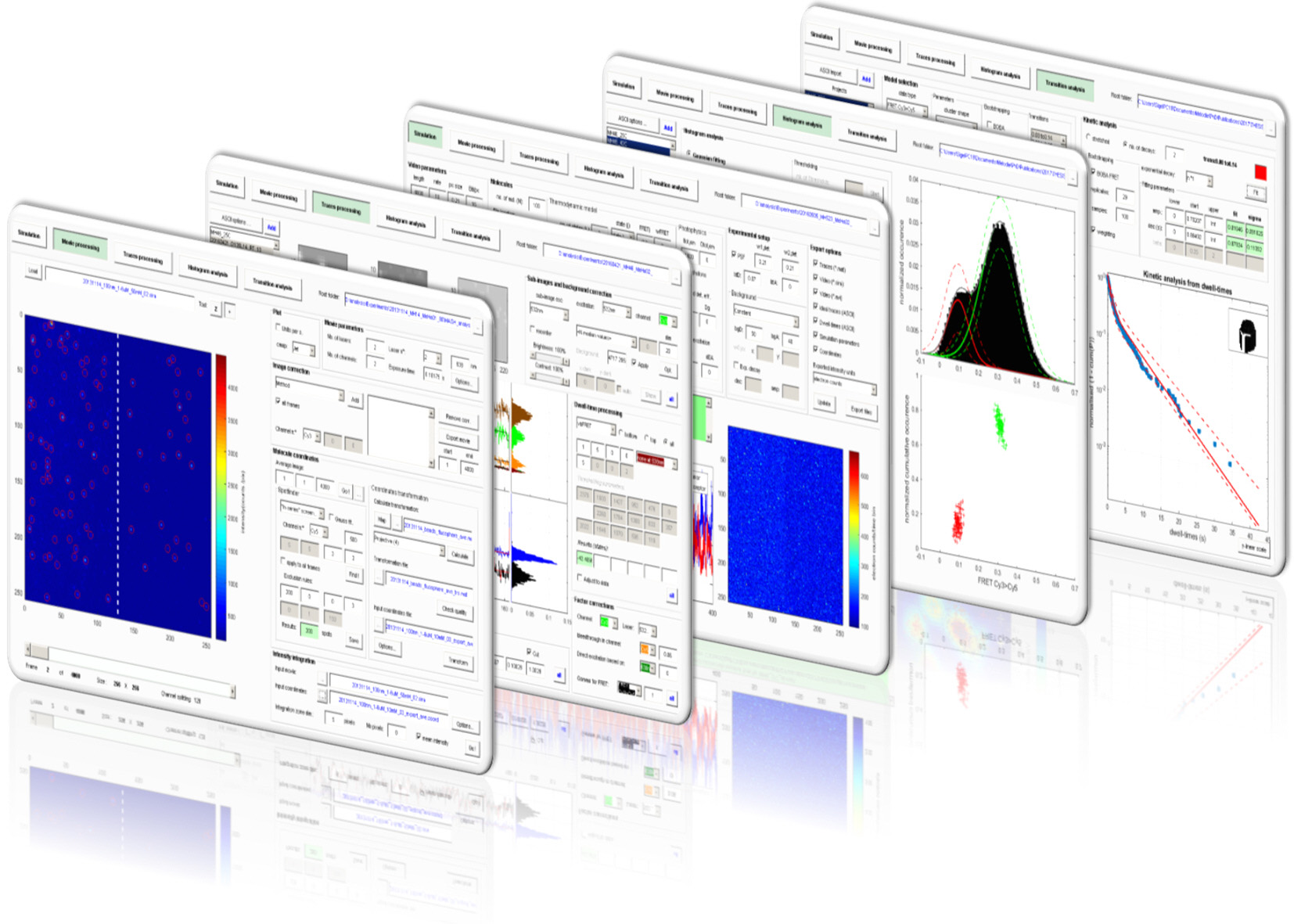
The MATLAB-based Multifunctional Analysis Software for Heterogeneous single molecule FRET data (MASH) facilitates sm-FRET data evaluation and simplifies the comparison of several existing algorithms. The graphical interface permits a complete data analysis with a choice of existing methods and allows to perform realistic simulations.
FRETraj is a Python module for predicting FRET efficiencies by calculating multiple accessible-contact volumes (multi-ACV) to estimate donor and acceptor dye dynamics. The package features a user-friendly PyMOL plugin for for FRET-assisted, integrative structural modeling. It interfaces with the LabelLib library for fast computation of ACVs. Specifically, FRETraj is designed to:
- plan (single-molecule) FRET experiments by optimizing labeling positions
- interpret FRET-based distance measurements on a biomolecule
- integrate FRET experiments with molecular dynamics simulations
The web-interfaced database for Metal Ions in Nucleic AcidS (MINAS) allows to search for metal ions in nucleic acid structures contained in PDB and NDB. It compiles the detailed information on innersphere, outersphere and larger coordination environment.
The MATLAB script for determination of Intrinsic STAbilities of RNA complexes (ISTAR) calculates the intrinsic affinity constants of M2+ ions to specific binding sites in nucleic acids from the NMR chemical shift values.
All software and scripts are freely available and accessible from the "software" section of the website.
Literature
M.C.A.S. Hadzic, R. K.O. Sigel*, and R. Börner*, in DNAzymes. Methods in Molecular Biology, 2022, (2439), 173-190.
doi https://doi.org/10.1007/978-1-0716-2047-2_12
F.D. Steffen, R.K.O. Sigel, R. Börner, Bioinformatics 2021.
M.C.A.S. Hadzic, R. Börner*, D. Kowerko, S.L.B. Koenig, and R. K.O. Sigel*, J. of Phys. Chem. B, 2018, epub, online.
doi:10.1021/acs.jpcb.7b12483.
R. Börner*, D. Kowerko, M. C. A. S. Hadzic, S. L. B. König, M. Ritter, Roland K. O. Sigel*, PlosOne, 2018, 13, e0195277.
doi:10.1371/journal.pone.0195277 .
R. Börner*, D. Kowerko*, H. G. Miserachs, M. F. Schaffer and R. K. O. Sigel*, Coord. Chem. Rev., 2016, 327-328, 123–142.
doi:10.1016/j.ccr.2016.06.002
M. C. A. S. Hadzic, D. Kowerko, R. Börner, S. Zelger-Paulus and R. K. O. Sigel*, Proc. SPIE, 2016, 9711, 971119.
doi:10.1117/12.2211191
A. Böttcher, D. Kowerko and R. K. O. Sigel*, Biophys. Chem., 2015, 202, 32–39.
doi:10.1016/j.bpc.2015.04.001
S. L. B. König, M. C. A. S. Hadzic, E. Fiorini, R. Börner, D. Kowerko, W. U. Blanckenhorn and R. K. O. Sigel*, PLoS One, 2013, 8, e84157.
doi:10.1371/journal.pone.0084157
M. C. Erat, J. Coles, C. Finazzo, B. Knobloch and R. K. Sigel*, Coord. Chem. Rev., 2012, 256, 279–288.
doi:10.1016/j.ccr.2011.08.009
J. Schnabl, P. Suter and R. K. O. Sigel*, Nucleic Acids Res., 2012, 40, D434-8.
doi:10.1093/nar/gkr920